Projects
High Throughput In-Incubator Fluorescence Microscope
As a part of my work as a researcher at the UC Santa Cruz Genomics Institute, I designed and developed a modular open source microscopy framework for creating live cell fluorescence imaging systems that can operate inside the hot and humid conditions of a cell culture incubator (37 degrees C/5% CO2 concentration). This work included designing and selecting optical microscopy components, fabricating custom motion systems that can operate safely inside of the incubator, and writing firmware and software and control systems that allow for the continuous operation of this system and the high-throughput capture of images without damaging live samples. To demonstrate the effectiveness of my system, I worked with biologists affiliated with my research group to conduct long term live cell imaging experiment with run times anywhere from 2 days to 2 weeks, with imaging intervals measured in minutes.
I have created versions of this system that can perform single and multi-color fluorescence, as well as having optional plate-scanning capabilities through the use of a custom three axis motion stage. As part of other collaborations (see below sections), I have also developed experiment-specific versions of the microsope meant for long-term in-incubator brightfield imaging.
As of January 2026, I have published our preprint for this system, with a journal submission in progress.
Figure from preprint showing the design of our system:
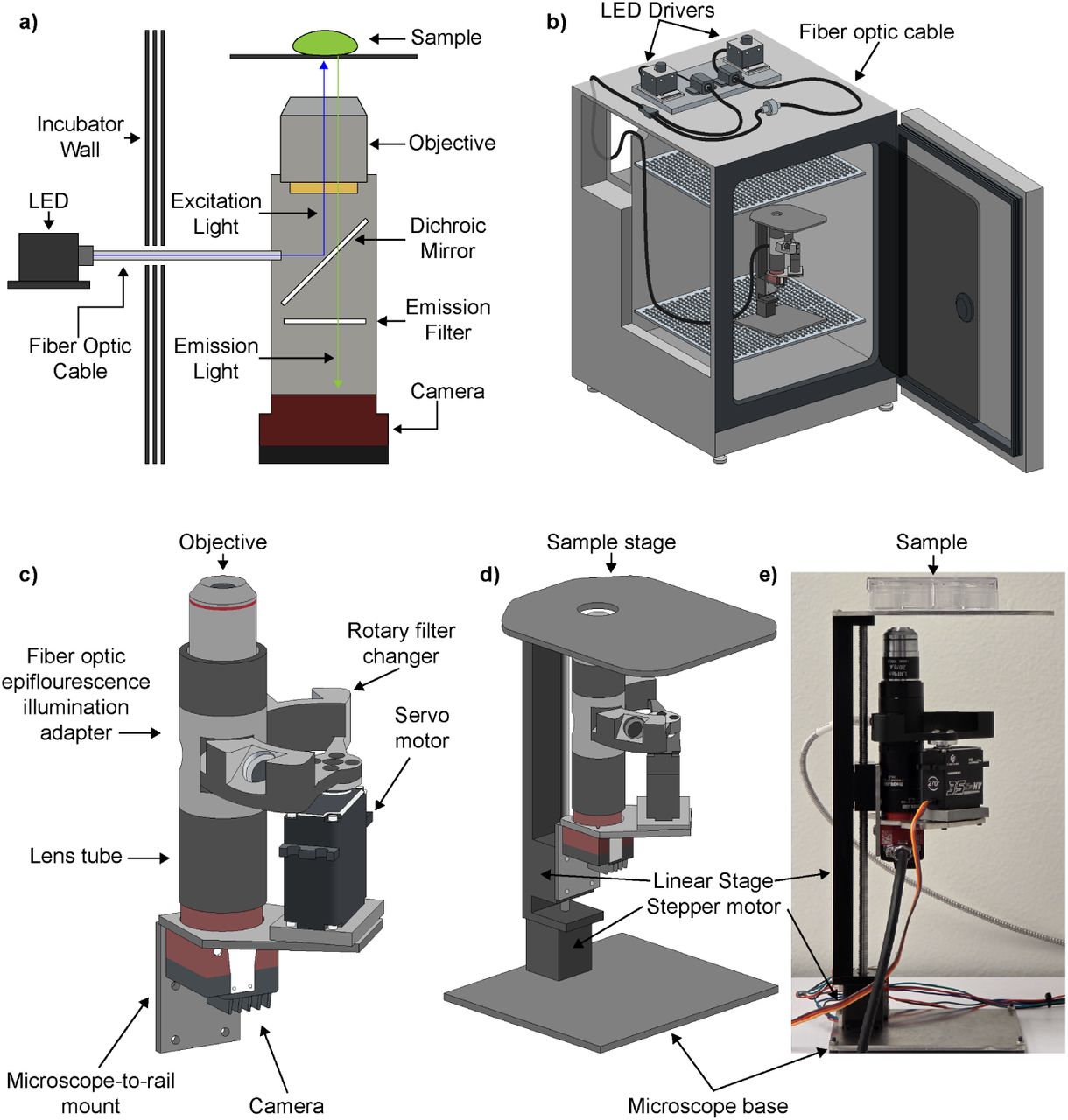
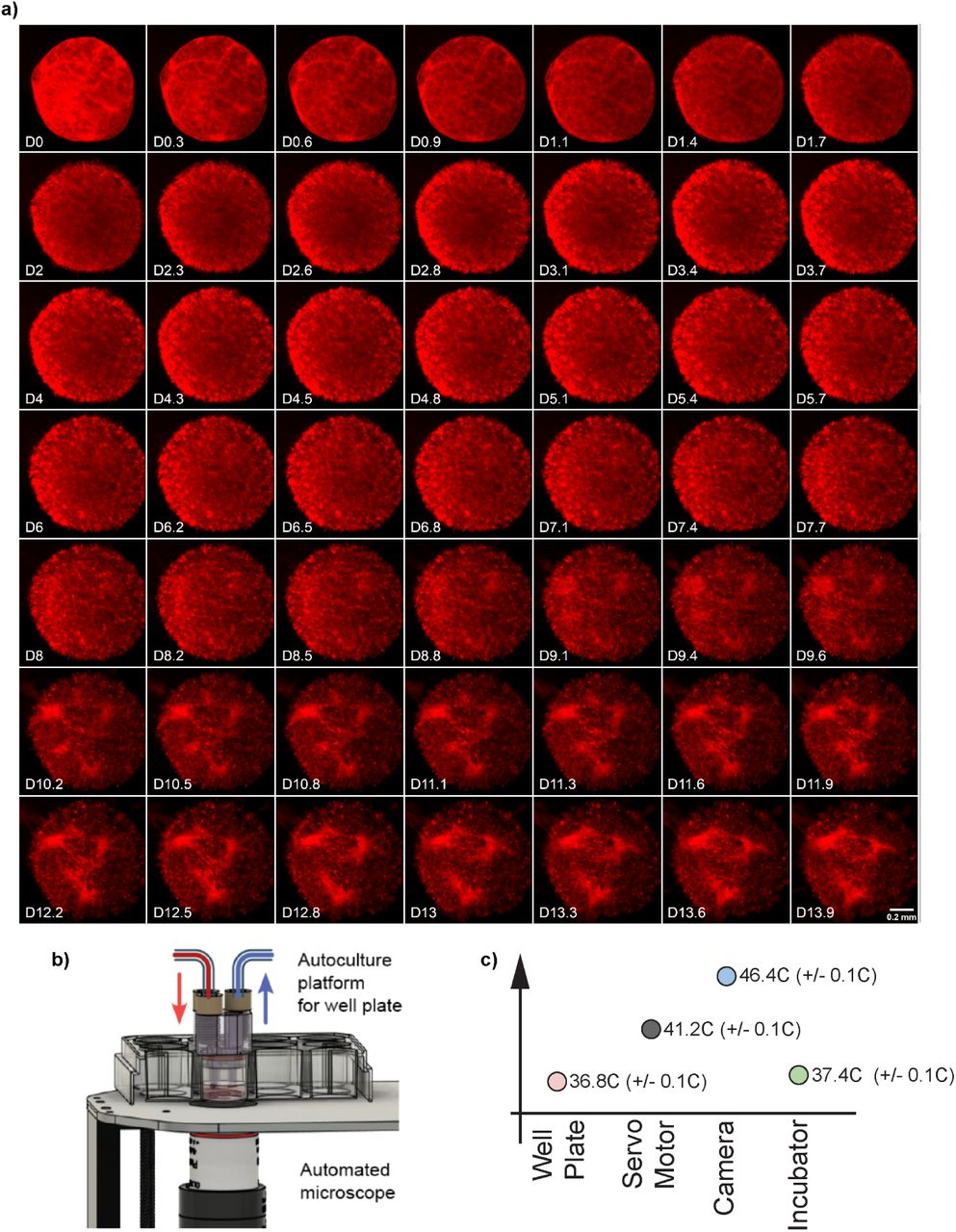
Figure from preprint demonstrating longitudinal capabilities of microscopy system, each sqaure in the grid is approximately 8 hours - images were taken every 2 minutes, but this figure shows an abridged data set for visual clarity:
Remote Microscopy for Education
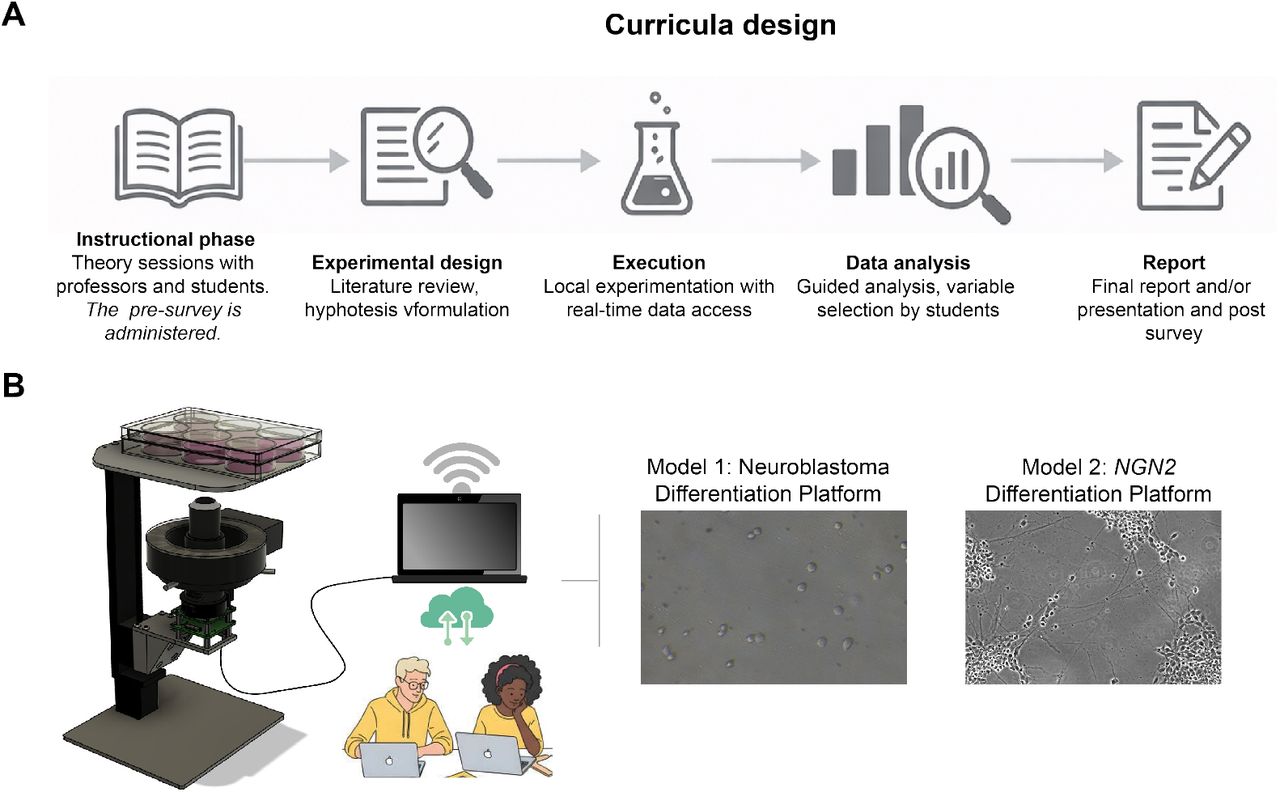
In collaboration with the Live Cell Biotechnology Discovery Lab at the UC Santa Cruz Genomics Institute, I have provided the use of a brightfield variant of the microscope described in the previous section for livestreamed science education experiments. This has taken the form of modifying and programming the microscope to capture brightfield images of cell cultures that have been chosen by the participating students and make the results accessible in near real-time online. We have collaborated with multiple schools in multiple countries as part of a broader initiative to make laboratory neuroscience more accessible in areas that don’t have the resources to perform these experiments themselves. We are still continuing to perform these experiments throught 2026.
My contribution to this work has been to provide and manage custom imaging equipment, software, and livestreaming. This work has resulted in contributions three publications so far.
Visual abstract for the most recent publication, of which I am the second author:
Seru-Otchi: Laboratory Protocol Training Game for Undergraduates
Undergraduate students looking to join a scientific laboratory often have to read intimidating and dense academic articles that are very difficult to derive knowledge from unless you have experience with reading them. This work, a collaboration between Dr. Victoria Ly, Dr. Jess Sevetson, and myself, looks to help lower the barrier to entry for undergraduates joining a lab by supplementing the academic materials with an interactive browser based experience that creates a interactive version of the paper used by Dr. Sevetson to teach new students cell culture.
This work and the study we did to validate it has resulted in a paper that been published in Heliyon, a Cell Press journal.
Gamifying cell culture training: The ‘Seru-Otchi’ experience for undergraduates

PolyPhy: 3D Printing Slime Mold Structures
The Physarum polycephalum slime mold is an acellular organism that can solve shortest paths problems due to its optimal foraging problems. Because of this, Dr. Oskar Elek, in collaboration with cosmologists and data visualization experts, created a Monte Carlo-based simulation software to use the behaviors of the slime mold to approximate the shape of the ‘cosmic web’, a gigantic network of gas and dark matter that can tell us a lot about the formation of galaxies and the structure of the universe.
My contribution to this project has been to take the structures created by the simulation software, PolyPhy (previously called PolyPhorm) and turn them into 3D printable meshes. These meshes should retain some of the topologically optimal properties of the slime mold, and are visually compelling bio-inspired objects. With my pipeline, any watertight 3D mesh can be ‘slimed’ and turned into a representation of itself created out of dense networks of scaffolding structures.
The earlier versions of this project formed the basis for my bachelor’s and master’s theses. More recently, this work has resulted in two poster presentations at the ACM Symposium on Computational Fabrication (2022,2023), with a paper focused on the more technical aspects of the project coming soon.

Alternate Controllers for Team Liquid
After winning the first edition of major eSports organization Team Liquid’s hackahthon, Liquid Hacks, my team and I were invited to participate in two pieces of video content for their YouTube channel. The first video featured the project we created for the hackathon, which turned a Nerf blaster into a controller for the video game Valorant. We shipped our controller to the Netherlands for their professional Valorant team to try out, and the video is a compilation of their attempts and our reactions.
For the second video, Team Liquid flew my team and I out to Los Angeles to spend a weekend building an alternate controller for League of Legends based on a specific character’s abilities. This controller was then used by one of their professional players, a world champion who is one of the best at that character on the planet.
This contract work has resulted in two published pieces of video content that feature my engineering work prominently.
Can CoreJJ play with an IRL Thresh Hook?!, 2022
Liquid Play VALORANT In VR ?! ft. Liquid Hacks, 2021
Selected Hackathon Projects
I participated in many hackathons as an undergraduate as excuses to learn new things and as a creative outlet - if the project idea didn’t make me laugh, I probably wasn’t interested. Here are some of my personal favorite projects that I was a part of. My full Devpost profile can be found here.
Archie, Warmer of Snacks - Winner, Best Hardware Hack, CruzHacks 2021
R.A.N.G.E. - Winner, 1st Overall, LiquidHacks 2020
owie, - Winner, “Well, it’s not rocket science.” Prize, To the Moon and Hack 2020
